Tracking down leafcutter ants: Mission impossible?
You’re in the rainforest. Its 30 degrees plus hot and humid, you’re sweating from every pore, and you have a very specific goal: to find leafcutter ants. On first appearances this seems a lot like finding a needle in a haystack…
Before you head out you need to grab a selection of digging tools (unexpected use for a dessert spoon), something to cut with (large branches to thread-like roots), containers of different shapes and sizes, and a medley of problem-solving tools (forceps, tape, bin bags…). Plus a good supply of water, long sleeves, wellies, and don’t forget the bug spray – everything that can bite you, will.
And then you’re off.
The basic strategy is ‘search for small moving things, carrying pieces of leaf-litter’: yep, that’s right, you’re looking for leaves among leaf litter on the forest floor.
I imagine it’s a bit like tracking a bear (I’ve never done it) except much, much smaller. Or the fieldwork equivalent of those police stakeouts in the movies… but with fewer rounds of coffee and doughnuts: you prop yourself up, you wait, and you watch.
Very carefully.
When you spot something promising the fun really starts, trying to follow the twists and turns of the ant’s foraging path all the way back to its colony entrance. It’s not uncommon that you’ll step on the ant’s odour trail before you spot the ant, leaving the ant running in circles and you back at square one. If you’re lucky the ant you spot doesn’t drop so deep into the leaf litter that you lose it, the colony is close, and the entrance is obvious – in reality, one out of these three is good going.
A few cardinal rules – e.g. don’t dig in the rain – and a lot of patience later, you hit upon an ant colony.
The attine ants (leafcutters and their relatives) share a novel way of getting food: they are all fungus-farmers. They vary in size from species of a few millimetres, to others where the large soldiers can be an inch or three (not to mention the monster Queens in the big leafcutter Atta species).
They vary in the places they build their nests and how big a colony is. There are the ones you can’t miss: the large leafcutter species, possibly familiar from natural history documentaries. You don’t have to hunt so hard for these but they can take hours of digging to collect, while defending yourself from the bites of angry soldier ants. But many other species you have to hunt down – just finding a colony can take hours. Some you find in cavities in the banks along rainforest creeks. Some have 10-plus separate chambers, up to a metre underground. Others hang their colonies in a kind of net, under fallen trees or rocks. And make no mistake, a longer hunt or a smaller ant species make little difference to how long you have to dig or how dirty you get in the process.
So why are we interested?
No doubt about it, fungus-farming ants are cool (they were developing agriculture before the great apes evolved). But these ants are also one of the most useful systems we have to try and better understand how mutualism – the name for cooperation that happens between two or more different species – works in nature.
Fungus farming ants, as with any successful farmers, need to both tend their crop and protect it from natural enemies. This led to three principle players as this mutualism developed: the ants, the fungus they cultivate as a crop, and unique bacteria that grow on the ant cuticles while producing antibiotics that the ants use as a pesticide on their fungus garden.
Mutualism is important in unexpected places: crop plants that fix nitrogen from the soil in their roots, coral bleaching or the conservation implications of this phenomenon, and human diseases affected by gut bacteria all involve mutualisms (or mutualism breaking down).
We’re interested in mutualism, how it evolves, whether and how these different species have, over evolutionary time, developed adaptations that increase the stability of this mutualism. We’re interested in the bacterial community that forms on the ants, and the antibiotics they produce. And much more besides.
Plus I defy anyone to hunt down a colony and not be taken aback by the complexity, curiosity, and unbelievable degree of organisational efficiency.
So, why are we interested in leaf cutter ants? My best answer is why on earth wouldn’t we be.
Tabitha Innocent is a Marie Curie postdoctoral research fellow at the Centre for Social Evolution, University of Copenhagen; CSE researchers are collaborating with the Hutchings group on this work.
http://socialevolution.ku.dk
http://www.uea.ac.uk/leafcutter-ants
Follow @Ant_ibiotics
Royal Society summer exhibition 2014
From Monday 30th June to Sunday 6th July 2014 me (Matt Hutchings) and my colleagues will be presenting a “Leafcutter ants and their antibiotics” exhibit at the Royal Society Summer Exhibition and I’ll be hijacking microbelog to post updates on our preparations and on the event itself. This is open to everyone so if you’re in London on any of the following dates please come along!
Mon 30 June
Exhibition is open to school groups 11am – 5pm
(followed by public evening event 6pm – 10pm)
Tues 1 July
Exhibition is open to the public 10am – 9pm
Wed 2 July
Exhibition is open to the public 10am – 5pm
(followed by VIP soirée)
Thu 3 July
Exhibition is open to the public 10am – 5pm
(followed by VIP soirée)
Fri 4 July
Exhibition is open to the public 10am – 8pm
Sat 5 July
Exhibition is open to the public 10am – 6pm
Sun 6 July
Exhibition is open to the public 10am – 6pm
Breast really is best
Microbiomes are the beneficial microbial communities associated with plants and animals. We all have them and they influence everything about their host from development and fitness to reproduction. Needless to say, microbes are a huge driving force in the evolution of higher organisms.
The microbiome that most concerns humans is the one in our guts, and which primes our immune systems, helps us digest our food and protects us against infection. We acquire some of our ‘good bacteria’ from our mums when we pass through the birth canal but many of the bacteria found in our guts are obligate anaerobes which are killed by exposure to air. So how do they get from mother and child?
Recently it has been suggested that bacteria may actually migrate from the mothers gut to the mammary glands and be transmitted directly to babies through her breast milk. A paper just published in Environmental Microbiology provides the first direct evidence for this ‘mother-neonate direct transfer’ model.
The authors used culture dependent and independent methods to examine the microbes present in the faeces (poo to you) of mums and their newborn babies (neonates) and in the mum’s breast milk. They found the same species in all three samples for mother-baby pairs and the highest evidence for direct transfer was for strains of bifidobacteria, the Y shaped “good bacteria” that are added to probiotic drinks and yoghurts.
Although this study was carried out in collaboration with Nestle (who have a big line in probiotics) it is backed up by other recent studies and it certainly makes a lot of sense. Exactly how the bacteria get from the mother’s gut to the breast milk remains mysterious but expect rapid progress to be made in figuring this out. And remember, breast really is best.
I’m lichen that !
 I’ve been fascinated by lichens since I studied them on a biology field trip to Pembrokeshire many moons ago (geeky, I know). Although they look like weird plants that only grow in barren rocky landscapes they are actually microrganisms. Or rather, they are a collection of symbiotic microorganisms; a fungus and a cyanobacterium or green algae colonised by lots of other bacteria. They have been around for 600M years!!
I’ve been fascinated by lichens since I studied them on a biology field trip to Pembrokeshire many moons ago (geeky, I know). Although they look like weird plants that only grow in barren rocky landscapes they are actually microrganisms. Or rather, they are a collection of symbiotic microorganisms; a fungus and a cyanobacterium or green algae colonised by lots of other bacteria. They have been around for 600M years!!
A new study reportsthat lichens make a class of antibiotic called pederins only previously seen in bacterial symbionts of Paedurus beetles. The researchers used metagenomics (sequencing all of the DNA from all of the microorganisms in the lichen at the same time) and then looked at this big mix of DNA to see if they could find any genes involved in making antibiotics. Voila, they found a gene cluster for a pederin-type compound and then purified it, solved the structure and called it nosperin. This work is exciting because it suggests some antibiotics may be specific to symbionts, even when they are distantly related (in this case, beetles and lichens)!
It also suggests that the lichen symbiosis, like others we have written about (e.g. fungus growing ants and marine sponges), could be a source of novel antibiotics. So lichens might provide us with yet another untapped and unusual niche for natural product discovery which could eventually mean new antibiotics and new anticancer compounds in the clinic (hurray!). For me at least, this will be easier than sampling the marine environment, because we don’t have any submarines where I work.
Matt Hutchings
@MattHutchings10
Clues to the evolution of disease?
What are a group of genes that help Mycobacterium tuberculosis infect mammalian cells doing in a harmless soil bacterium? Dr Paul Hoskisson explains his recent research, which is helping us understand the evolutionary roots of disease.
 Scientists invest a lot of time and resources in trying to understand how bacteria cause disease. Generally, this involves studying a particular gene, or group of genes, in a pathogenic (disease-causing) bacterium to see what function it has during the infection process. In my laboratory, we have been trying to understand the evolutionary processes that cause these genes to develop.
Scientists invest a lot of time and resources in trying to understand how bacteria cause disease. Generally, this involves studying a particular gene, or group of genes, in a pathogenic (disease-causing) bacterium to see what function it has during the infection process. In my laboratory, we have been trying to understand the evolutionary processes that cause these genes to develop.
Bacteria need some specialist skills if they’re going to cause disease: they must avoid being destroyed by the immune system, enter and multiply within host cells, and produce toxins. Bacteria have been around on Earth a lot longer than humans, which suggests that the genes needed to cause disease in humans have either evolved recently and rapidly (in evolutionary terms) or been co-opted from existing genes in harmless bacteria.
We noticed that the soil bacterium Streptomyces coelicolor has a cluster of genes that are very similar to pathogenicity genes present in Mycobacterium tuberculosis, the causative agent of tuberculosis. Other work has shown that these genes, known as mce (mammalian cell entry) genes, are used by M. tuberculosis to colonise and survive inside human immune cells called macrophages. We disrupted these genes in S. coelicolor to see what happened.
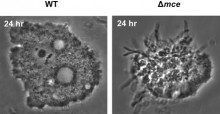
As the mce genes are used by M. tuberculosis to infect and survive in macrophages, we wanted to see what happened to the mce-deficient S. coelicolor mutants under similar conditions. However, because S. coelicolor lives in the soil, it rarely encounters any immune cells, so we had to get creative.
Rather than use immune cells, we used amoebae. This might not seem like an obvious choice, but these single-celled organisms are eukaryotic, just like immune cells, and engulf bacteria in a similar way. We got a strange result when we fed the S. coelicolor mce mutants to the amoebae: the amoebae all died (see image above). This effect has been seen in M. tuberculosis as well – mce mutants are hyper-virulent and much better at killing immune cells than regular M. tuberculosis. This is counterintuitive, but while the mutant M. tuberculosis strain may be excellent at killing macrophages, it is unable to cause disease in a whole animal – presumably because of the presence of the complete immune system, which is much more complex.
We did a little detective work and discovered that the mce genes encode a transporter that bacteria use to take up sterol (a type of fat), which they can use as a source of food, from the environment. It is thought that M. tuberculosis uses this transporter to take up sterols because other nutrients are hard to find inside macrophages.
One location where Streptomyces encounters sterols in the environment is around the roots of plants, an environment known as the rhizosphere. We showed that the S. coelicolor mce mutant is poorer at colonising plant roots than the wild-type S. coelicolor, which suggests that sterols are an important carbon source for these bacteria.
We suggest that as these bacterial species adapted to their particular environment – soil in the case of the Streptomyces and human immune cells for M. tuberculosis – their respective mce genes evolved to operate under the different conditions. Understanding how the selection process has led to the changes in function are essential to our appreciation of the evolution of virulence in bacteria and could significantly aid our understanding of the emergence of pathogenic bacterial strains.
Paul A Hoskisson is a lecturer in microbiology at the University of Strathclyde. His research focuses on antibiotic-producing actinomycetes and emerging actinomycete pathogens.
Image adapted from the original article published in Scientific Reports article under the Open Access license.
Clark LC et al. Mammalian cell entry genes in Streptomyces may provide clues to the evolution of bacterial virulence. Sci Rep 2013;3:1109. doi:10.1038/srep01109
No barrier to cell division
 Bacterial cells are surrounded by a highly cross-linked cell wall that has to be constantly broken down and remade as the cells get bigger. Since most bacteria reproduce by dividing the cell into two they also have to build a new bit of cell wall in the middle of the cell to make two daughter cells. In bacteria the FtsZ protein forms a Z ring to mark this site of cell division and all the other cell division proteins assemble on this Z ring and remodel the cell wall.
Bacterial cells are surrounded by a highly cross-linked cell wall that has to be constantly broken down and remade as the cells get bigger. Since most bacteria reproduce by dividing the cell into two they also have to build a new bit of cell wall in the middle of the cell to make two daughter cells. In bacteria the FtsZ protein forms a Z ring to mark this site of cell division and all the other cell division proteins assemble on this Z ring and remodel the cell wall.
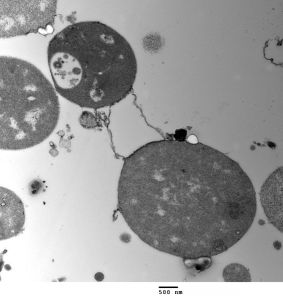
Despite this, it has been known for a long time that some bacteria can exist as L forms which have lost their cell wall. Even more amazing, these L-form bacteria don’t need FtsZ to divide and instead they just bud off membrane vesicles, some of which contain DNA and enough proteins to form a new cell.
Jeff Errington’s group in Newcastle recently made stable L forms of a bacterium called Bacillus subtilis, effectively reversing 3 billion years of evolution . These bacteria can live without a wall or FtsZ but they still need a cell membrane to contain the cell contents and in another groundbreaking paper Jeff’s group have shown that simply increasing the amount of membrane the cell makes is enough to make these L-forms divide.
The beauty of these L forms is that they help us to understand how the first living cells, reproduced themselves around 4 billion years ago. In the absence of a cell wall and a complicated cell division machinery they simply increased their surface area to volume ratio in order to propagate. And that, frankly, is amazing.
Mercier R, Kawai Y and Errington J (2013). Excess membrane synthesis drives a primitive mode of cell proliferation. Cell 152 997-1007.
Image credit: The Red Lexicon on Wikimedia Commons
Posted by Matt Hutchings
Animals in a bacterial world
 This isn’t a research blog as such, just an attempt to get everyone to read a new perspective article entitled “Animals in a bacterial world”. Here at microbelog we’ve been banging on about how important symbioses between bacteria and eukaryotes are for a quite a while now. So imagine our excitement to read this article in the latest issue of the Proceedings of the National Academy of Sciences USA. Although slightly evangelical (or as one of my evolutionary biologist colleagues put it “messianic”), this is well worth a read. It’s also good to see Carl Woese and Lynn Margulis, both of whom sadly passed away recently, getting the credit they deserve. Facts like one third of human DNA is bacterial in origin is the kind of thing that gets us very excited! This is a call to arms and it’s certainly never been a more exciting time to be a microbiologist!
This isn’t a research blog as such, just an attempt to get everyone to read a new perspective article entitled “Animals in a bacterial world”. Here at microbelog we’ve been banging on about how important symbioses between bacteria and eukaryotes are for a quite a while now. So imagine our excitement to read this article in the latest issue of the Proceedings of the National Academy of Sciences USA. Although slightly evangelical (or as one of my evolutionary biologist colleagues put it “messianic”), this is well worth a read. It’s also good to see Carl Woese and Lynn Margulis, both of whom sadly passed away recently, getting the credit they deserve. Facts like one third of human DNA is bacterial in origin is the kind of thing that gets us very excited! This is a call to arms and it’s certainly never been a more exciting time to be a microbiologist!
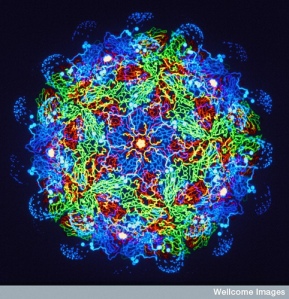
Image credit: Picture of a captive leaf-cutter ant colony taken by Andrew Smith, John Innes Centre, Norwich 2012.
The sting in the tail of antibiotic use

![]() Recently, I found a paper published in mBIO that describes how antibiotic use in farming is involved in the spread of resistance genes. In this case the work focuses on the humble honeybee (Apis mellifera). Since the 1950s, beekeepers in the USA have been using the antibiotic oxytetracycline – a ‘broad-spectrum’ antibiotic that kills most species of bacteria – to prevent infections that can cause ‘foul brood’, a disease that kills bee larvae. As you can imagine, using a single antibiotic for more than 50 years has led to some selective pressure. In this work, researchers from Yale University were investigating the prevalence of disease resistance in bee gut bacteria.
Recently, I found a paper published in mBIO that describes how antibiotic use in farming is involved in the spread of resistance genes. In this case the work focuses on the humble honeybee (Apis mellifera). Since the 1950s, beekeepers in the USA have been using the antibiotic oxytetracycline – a ‘broad-spectrum’ antibiotic that kills most species of bacteria – to prevent infections that can cause ‘foul brood’, a disease that kills bee larvae. As you can imagine, using a single antibiotic for more than 50 years has led to some selective pressure. In this work, researchers from Yale University were investigating the prevalence of disease resistance in bee gut bacteria.
This might seem like a strange place to look, but it actually has its advantages. Unlike the supremely complex ecosystem of the human gut microbiome, the bee’s is pretty simple, with eight species making up over 95% of the gut bacteria in adult worker bees. The small number of species and the knowledge of how hives have been treated allowed the researchers to monitor the impact of decades of antibiotic use.
Leaking leachate
![]() Sometimes you find a paper with a title so intriguing you just have to find out a little more about it. Recently, I came across a paper about ‘entombed pigs’, so how could I possibly ignore it? I learned a fair bit about animal-disease control methods in Asia and the use of quicklime to decompose corpses , a fairly standard weekend for me.
Sometimes you find a paper with a title so intriguing you just have to find out a little more about it. Recently, I came across a paper about ‘entombed pigs’, so how could I possibly ignore it? I learned a fair bit about animal-disease control methods in Asia and the use of quicklime to decompose corpses , a fairly standard weekend for me.
The work centres on foot-and-mouth disease (FMD), a viral infection of hoofed animals caused by Aphthovirus. It causes significant suffering in animals and has serious economic consequences: a 2001 outbreak is estimated to have cost the UK £8 billion.
Millions of infected animals were culled in South Korea in 2010/11, then buried (rather than burnt, as they are in the UK). The slaughtered animals were placed in five-metre-deep pits and covered with quicklime and copious amounts of soil to prevent the FMD spreading. Problem solved? Well, perhaps not.
Passing a camel through the eye of a needle?
 A recent study led by Professor Tracy Palmer at the University of Dundee discovered a new way by which bacteria can insert proteins into membranes, using two different transport machines. Here she gives us the lowdown on what it’s all about…
A recent study led by Professor Tracy Palmer at the University of Dundee discovered a new way by which bacteria can insert proteins into membranes, using two different transport machines. Here she gives us the lowdown on what it’s all about…
All cells are surrounded by lipid membranes; they keep the insides in, and the outsides out, but sometimes bacteria need to bring a molecule from the environment into the cell, or vice versa. The membrane itself is pretty much impermeable, so the cell uses specialised proteins to make channels in the membrane so they can control what moves from one side to the other.